Building a Mini RAG for a Homelab Help system
Learn how to build a Retrieval-Augmented Generation pipeline: paraphrase docs, chunk text with overlap, convert to embeddings using BAAI/bge-base-en-v1.5, index with FAISS, and serve with FastAPI + OpenAI/Mistral ... from data prep to query handling.
Last time, we covered off the hosting of a quantised model, this time we're going to dive into Retrieval-Augmented Generation or RAG which is basically a mechanism that combines a fast vector search of our own documents and information with the capabilities of an LLM (Large-Language Model) ... super important for most teams/orgs that have their own knowledge bases that have been developed over years and years.
For the purpose of this article we'll take a look at the process, some simple data processing and eventually throw our data over to an LLM to provide a suitable response based on the data from our vector store.
At a high level it's simply:
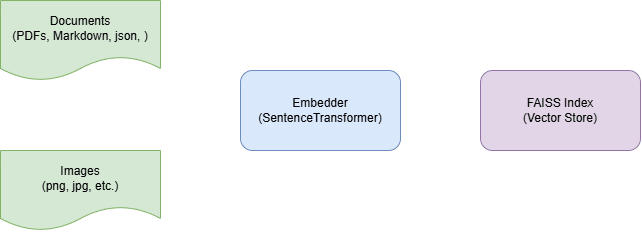
A Retrieval-Augmented Generation (RAG) pipeline typically begins by ingesting all of our raw content, including documents, images, PDFs and any other sources of knowledge, and chunking it into small, self-contained pieces. Each chunk is then passed through a pretrained SentenceTransformer or a similar model to produce a fixed-length embedding; a numerical vector that encodes the chunk's semantic meaning. Those embeddings are loaded into FAISS (Facebook AI Similarity Search), a highly optimised vector store designed to rapidly find nearest-neighbor vectors at scale.
At query time, we embed the user's question with the same model and submit it to FAISS, which immediately returns the top-K most relevant chunks. We concatenate those retrieved chunks into a context prompt alongside the original question and send the whole package to our language model (for example, via OpenAI’s Chat or Completion API). The LLM then generates an answer that's grounded in the retrieved text. By separating the concerns of storage (FAISS) and reasoning (the LLM), a RAG system lets us update our knowledge base without retraining or reloading large models and still deliver accurate, up-to-date responses.
Now we know what we're building let's dive in...
Data preparation
📂 Code Repository: Explore the complete code and configurations for this article series on GitHub.
To run this lab we need to some data... for this we'll leverage synthetic data in the form of json objects structured as follows, so the pipeline is reproducible without requiring private files:
{
{
"title": "Document Title",
"category": "Document Categorisation",
"tags": ["Array", "of", "tags", "associated", "with", "the", "document"],
"source": "document_file.md",
"last_updated": "2025-05-09",
"content": "Document content"
}To generate this we'll define a bunch of categories and apps and run something like:
def generate_docs():
docs = []
for category, titles in CATEGORIES.items():
for title in titles:
docs.append({
"title": title,
"category": category,
"tags": [category.lower()] + title.lower().split()[:2],
"content": generate_content(title, category),
"source": f"{category.lower()}-guide.md"
})
app_titles = generate_homelab_app_titles(n=70)
for title in app_titles:
docs.append({
"title": title,
"category": "HomelabApps",
"tags": ["homelabapps"] + title.lower().split()[:2],
"content": generate_content(title, "HomelabApps"),
"source": "apps-guide.md"
})
return sorted(docs, key=lambda d: d["title"])Now we have 100 documents, nicely structured and ready for processing, the first step of which is to paraphrase our content to ensure we inject some semantic diversity and noise into the chunks; this leads to a more robust, generalised RAG system once we embed and store them in FAISS.
To do this, we leverage the mistral-7B model to run the following:
MODEL_NAME = "mistralai/Mistral-7B-Instruct-v0.1"
PROMPT_TEMPLATE = """<s>[INST]
You are helping rewrite technical homelab Markdown articles.
Please paraphrase the following article:
- Keep the structure and Markdown formatting (e.g. headings, steps, bold)
- Change the wording and phrasing throughout
- Add 1-2 realistic gotchas or notes
- Preserve technical accuracy
Original:
{input}
Paraphrased:
[/INST]
"""
def load_seed_docs(path):
with open(path, "r", encoding="utf-8") as f:
return [json.loads(line) for line in f]
def main():
print("Loading model:", MODEL_NAME)
tokenizer = AutoTokenizer.from_pretrained(MODEL_NAME)
model = AutoModelForCausalLM.from_pretrained(
MODEL_NAME,
torch_dtype="auto",
device_map="auto"
)
paraphraser = pipeline(
"text-generation",
model=model,
tokenizer=tokenizer,
max_new_tokens=1024,
temperature=0.7
)
seed_docs = load_seed_docs("seed_documents/seed_docs.jsonl")
out_path = Path("paraphrased_documents/mixtral_paraphrased.jsonl")
out_path.parent.mkdir(parents=True, exist_ok=True)
with out_path.open("w", encoding="utf-8") as f:
for doc in tqdm(seed_docs, desc="Paraphrasing docs"):
prompt = PROMPT_TEMPLATE.format(input=doc["content"])
try:
result = paraphraser(prompt)[0]["generated_text"]
doc["paraphrased_content"] = result
f.write(json.dumps(doc) + "\n")
except Exception as e:
print(f"Error on {doc['title']}: {e}")
print("\nAll documents paraphrased!")Now we have our data ready for chunking and embedding into FAISS.
Chunking and embedding
Below is the core of our pipeline that takes our paraphrased documents, splits them into overlapping chunks, turns each chunk into a fixed-length embedding via BAAI/bge-base-en-v1.5, and finally writes everything into a FAISS index along with per-chunk metadata.
# CONFIG
PARAPHRASED_FILE = "paraphrased_documents/mixtral_paraphrased.jsonl"
CHUNK_SIZE, CHUNK_OVERLAP = 350, 100
EMBED_MODEL = "BAAI/bge-base-en-v1.5"
# Load the Sentence-Transformer embedder
from sentence_transformers import SentenceTransformer
embedder = SentenceTransformer(EMBED_MODEL)Here we point at our paraphrased JSONL, choose a 350-token chunk with 100-token overlap, and load BAAI’s BGE embedder for high-quality text vectors.
def chunk_text(text, size=CHUNK_SIZE, overlap=CHUNK_OVERLAP):
tokens = tokenizer.encode(text, add_special_tokens=False)
chunks = []
for i in range(0, len(tokens), size - overlap):
chunk = tokenizer.decode(tokens[i : i + size])
chunks.append(chunk)
if i + size >= len(tokens): break
return chunks
Here we split each document into passages of 350 tokens (≈250 words) and make them overlap by 100 tokens (≈75 words).
- Chunk size controls how much context lives in each vector: larger chunks carry more context so the LLM has a richer passage to pull from, but you get fewer "retrieval candidates"
- Overlap is what lets us "stitch" our chunks back together in meaning, so that any idea, entity, or fact that falls right at the end of one chunk still appears at the start of the next. This means we preserve the continuity of response, reduce retrieval errors and smooth out the transitions when we pass it to the LLM
Choosing the right values:
- If you need finer-grained retrieval (e.g. very specific paragraphs), reduce size and keep overlap around 20–30% of that window
- If you want broader context in each hit (e.g. entire sections), increase size and proportionally bump up overlap so you still capture boundary ideas
all_embeddings, metadata = [], []
for doc in docs:
for i, chunk in enumerate(chunk_text(doc["paraphrased_content"])):
prefixed = f"Represent this passage for retrieval: {chunk}"
emb = embedder.encode([prefixed])[0]
metadata.append({...})
all_embeddings.append(emb)
We prepend each chunk with a tiny "retrieval" instruction (e.g. "Represent this passage for retrieval: …"), then feed them in batches into our SentenceTransformer to get fixed-length vectors. For each vector we keep only the essentials (document title, source, category and chunk index) so FAISS can return both the embedding and just enough metadata to rehydrate the passage for answering queries.
import faiss, numpy as np
index = faiss.IndexFlatL2(all_embeddings[0].shape[0])
index.add(np.vstack(all_embeddings).astype("float32"))
faiss.write_index(index, "embedded_chunks/index.faiss")
Lastly, we use FAISS's IndexFlatL2, which simply keeps all of our chunk embeddings in a single, in-memory "flat" structure and computes exact nearest-neighbour searches using Euclidean (L2) distance. Because it doesn't approximate or quantise, we get precise similarity rankings at the cost of a linear scan, but for a few thousand chunks this remains blazing fast. Once built, we serialise the index to disk so that on startup we can instantly reload it and jump straight into similarity lookups without re-embedding or rebuilding anything.
With this in place, at query-time we can embed the user question, perform a nearest-neighbours lookup in FAISS, pull back the top k chunks, and feed their texts (plus our prompt formatting) into our LLM to produce a grounded answer.
What we end up with here is an index.faiss file and a metadata.json file that we can now leverage as part of our RAG service.
Serving the RAG
Now we have our index.faiss and metadata.json file we can wrap this in a FastAPI service and leverage an LLM to process the chunks and produce something useful based on our docs and vector store.
In essence, what we'll do is:
- Embed & Tokenise the Query - We take the user's question, prepend our "represent this passage" instruction, and run it through our Sentence-Transformer embedder. That turns it into a fixed-length vector in the same space as our document chunks
- FAISS Nearest-Neighbor Search - FAISS then finds the top-K chunk vectors whose embeddings are closest to the query vector. Those chunks (titles, source paths, and text) are the most relevant snippets from our knowledge base
- Contextual Generation with an LLM - Finally, we stitch those retrieved chunks into a single "context" block and feed that, along with the original question, into our LLM (e.g. GPT-3.5 or Mistral). The model composes a coherent answer strictly based on the provided context
# Load the sentence-transformer we'll use for vectorizing queries
embedder = SentenceTransformer(EMBED_MODEL)
# Load our precomputed FAISS index…
index = faiss.read_index(INDEX_PATH)
# …and the accompanying metadata (titles, sources, original text)
with open(METADATA_PATH) as f:
metadata = json.load(f)
Before we can retrieve or generate anything, we need the embedder, the FAISS index, and the chunk metadata in memory. Loading them once at startup maximises throughput.
def search_faiss(query: str, top_k: int = TOP_K):
# Embed the query with the same “prefix” we used when indexing
prompt = f"Represent this passage for retrieval: {query}"
emb = embedder.encode([prompt], convert_to_numpy=True).astype("float32")
# Find the top_k closest embeddings in FAISS
_, idx = index.search(emb, top_k)
# Map those indices back to our metadata entries
return [metadata[i] for i in idx[0]]
Next we build a mini function that we use to search the FAISS index by converting the user's question into the same embedding space as the document chunks, then use FAISS's nearest-neighbor search to pull back the most relevant chunks of text.
def generate_via_openai(context: str, question: str) -> str:
resp = openai.chat.completions.create(
model="gpt-3.5-turbo",
messages=[
{"role":"system","content":"You are an expert assistant answering homelab questions."},
{"role":"user","content":f"Context:\n{context}\n\nQuestion: {question}"}
],
max_tokens=512, temperature=0.7
)
return resp.choices[0].message.content.strip()Next we take these chunks and feed them into an LLM to generate a reponse, in this case we'll leverage OpenAI to generate a response to the question "How do I setup pi-hole in my homelab?" like:
To set up Pi-hole in your homelab, follow these steps:
## Setting up Pi-hole in your Home Network
### Introduction
Pi-hole is a free and open-source software that allows you to manage your DNS server on a Raspberry Pi. This guide will walk you through the process of setting up Pi-hole in your home network.
### Prerequisites
Before you begin, ensure you have the following:
- A Raspberry Pi
- A microSD card with the latest version of Raspbian installed
- A power supply for your Raspberry Pi
- A network connection for your Raspberry Pi
### Installation Steps
1. Connect your Raspberry Pi to your network
2. Install Raspbian on your microSD card
3. Update your Raspberry Pi's software
4. Install Pi-hole
### Configuration Tips
- Set up secure credentials for your Pi-hole account
- Expose only necessary ports on your Raspberry Pi
- Use persistent volumes to store your Pi-hole configuration files
_Last Updated: 2025-05-03_
Note: Make sure to regularly update Pi-hole to ensure smooth operation and take advantage of new features or security patches.To generate this, we leveraged chunks from 2 different sources:
- Installing Pi-hole in your homelab (apps-guide.md)
- Running Pi-hole as a DNS filter (networking-guide.md)
With multiple chunks coming from each document... we have a response driven by our RAG ... this is pretty cool 😎
The core RAG plumbing, chunking, embedding and vector search, is surprisingly straightforward. The real heavy-lifting comes from parsing all your raw sources (PDFs, images, slides, docs) into clean, uniform text ready for chunking.
Next time
Up next, we'll walk through how to put our new RAG service through its paces: we'll build a small gold-standard QA suite to validate both retrieval accuracy and answer quality, set up automated metrics i.e. precision/recall for FAISS, F1 or ROUGE for the LLM's output, and establish regression tests so every change proves its value.
That hands-on evaluation will be the foundation for refining chunking, prompt design, and overall system robustness in the follow-on article.
